Phototrophic consortia
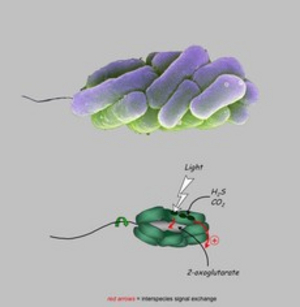
Phototrophic consortia consist of a fixed number of green sulfur bacteria epibionts surrounding a central colorless rod-shaped betaproteobacterium. Our group established the first laboratory cultures of phototrophic consortia that represent the only laboratory model system currently available for the study of highly specific interactions between nonrelated bacteria. Both partners divide in a highly coordinated manner. The entire consortium swims towards the light (scotophobic response), which the epibionts need for anoxygenic photosynthesis, whereas the central bacterium provides the necessary motility. The cells form specific cell-cell-adhesion structures and exchange multiple signals. Recent ultrastructural analyses have provided evidence for intracellular sorting of antenna structures in the epibiont cells, which occurs exclusively when they are in the symbiotic state. "Symbiosis genes" of the epibiont were identified by genomic approaches. Putative large exoproteins and a putative protein with a RTX toxin-type ß-roll could be identified. So far, such genes have not been found in any of the free living green sulfur bacteria, indicating a specific role in the interaction. Pulse-labeling and nanoSIMS analysis showed a physiological coupling of the phototrophic and heterotrophic partner by exchange of carbon and nitrogen compounds. Sincle cell genomic analysis of the bacteria constituting different types of phototrophic consortia indicate a long-term co-evolution of the partners. Lastly, laboratory cultures of phototrophic consortia also represent valuable model system for establishing suitable cryoconservation techniques for complex microbiomes.
Selected references:
- Liu, Z., Müller, J., Li, T., Alvey, R.M., Vogl, K., Frigaard, N.U., Rockwell, N.C. Boyd, E.S., Tomsho, L.P., Schuster, S.C., Henke, P., Rohde, M., Overmann, J., Bryant, D.A. (2013) Genomic analysis reveals key aspects of prokaryotic symbiosis in the phototrophic consortium “Chlorochromatium aggregatum”. Genome Biol. 14: R127.
- Müller, J., Overmann, J. (2011) Close interspecies interactions between prokaryotes from sulfureous environments. Front. Microbiol. 2: 146
- Wenter, R., Hütz, K., Dibbern, D., Li, T., Reisinger, V., Plöscher, M., Eichacker, L., Eddie, B., Hanson, T., Bryant, D.A., Overmann, J., (2010). Expression-based identification of genetic determinants of the bacterial symbiosis “Chlorochromatium aggregatum”. Environ. Microbiol. 12: 2259-2276
Autotroph-heterotroph interactions
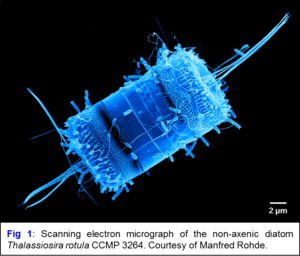
The beneficial effect of heterotrophic bacteria on the growth of algae and cyanobacteria has been observed by phycologists decades ago. Many phototrophs are auxotrophic for the synthesis of vitamins that are provided by their bacterial partners, which in turn benefit from the released exudates. One example is the diatom Thalassiosira rotula, which represents one of the most prominent cosmopolitan algae, but grows poorly under axenic conditions. In coculture with the dinoflagellate Prorocentrum minima, the bacterium Dinoroseobacter shibae shows a „Jekyll and Hyde“ behavior shifting from mutualism to pathogenicity during senescence of algal cultures. We identified a conjugative ‚killer plasmid‘ that is responsible for causing the morbid life style and are currently investigating its mode of action. We also systematically investigate the community composition of non-axenic microalgae and cyanobacteria with a new 16S-ITS high-throughput sequencing approach and metagenome sequencing in order to identify the key player(s) of the communities, using the growing cyanobacteria collection at the DSMZ. Promising strains are isolated and the interaction of model organisms is investigated to understand the specific metabolic interplay between the autotrophic and heterotrophic partners. This will not only improve our understanding of the physiological basis of interactions in cyanobacterial co-cultures, but also will add value to the cyanobacteria collection and guide subsequent attempts to obtain pure (axenic) cultures.
Projects
- Project A5, TRR51-Roseobacter
- Project A7, TRR51-Roseobacter
Selected References
- Segev E, Wyche TP, Kim KH, Petersen J, Ellebrandt C, Vlamakis H, et al. (2016). Dynamic metabolic exchange governs a marine algal-bacterial interaction. eLIFE 5: e17473.
- Wang H, Tomasch J, Michael V, Bhuju S, Jarek M, Petersen J, et al. (2015). Identification of genetic modules mediating the Jekyll and Hyde interaction of Dinoroseobacter shibaewith the dinoflagellate Prorocentrum minimum. Front. Microbiol. 6: 1262.
- Frank O, Michael V, Päuker O, Boedeker C, Jogler C, Rohde M, Petersen J (2015). Plasmid curing and the loss of grip - The 65-kb replicon of Phaeobacter inhibens DSM 17395 is required for biofilm formation, motility and the colonization of marine algae. Syst. Appl. Microbiol. 38: 120–127.
Interactions in heterotrophic co-cultures
In many environments, bacterial cells occur in close proximity to each other. The results of network analyses of complex microbial communities suggest that many heterotrophic bacteria interact under natural conditions. In addition, earlier results of our group showed that supplementation by bacterial signal compounds (homoserine lactones, cAMP) can significantly increase the growth and cultivation success of otherwise non-cultured bacteria. We systematically assess co-cultures of heterotrophic bacteria which are established in high-throughput from soil or aquatic samples and determine particular bacterial species which co-occur much more frequently than can be expected under random distribution. By this approach, potential keystone species can be identified and the co-cultures obtained permit the study of the physiological basis underlying specific bacteria-bacteria interactions.
Selected References
- Koch C, Huber KJ, Bunk B, Overmann J, Harnisch F (2019) Trophic networks improve the performance of microbial anodes treating wastewater. NPJ Biofilms Microbiomes 5, 27
- Overmann J, Abt B, Sikorski J (2017) Present and future of cultivating bacteria. Annu Rev Microbiol 71, 711–730
- Overmann J (2005) Chemotaxis and behavioural physiology of not-yet-cultivated microbes. Methods in Enzymology Vol. 397, Chapter II.8, p. 133-147. Elsevier, San Diego, USA

